Let’s write a function for our penguin species pickerInput that we can use in place of these two, rather long, chunks of code.
EDS 296: Part 6.1
Functions
Writing functions
Functions have many benefits and can improve your code base, particularly as your app grows in complexity
Learning Objectives - Functions
By the end of this section, you should:
understand the benefits of turning UI elements and server logic into functions
know where to write / save your functions
successfully turn a repeated input into a function
successfully turn a piece of server logic into a function
Why write functions?
Functions are useful for a wide variety of reasons. Most notably:
Where do I store my function(s)?
Importantly, functions can live outside of your app file(s) (i.e. app.R or ui.R / server.R / global.R), helping you to break up and streamline your code. Hadley Wickham recommends creating a folder called /R inside your app’s directory (e.g. ~/myApp/R/...) and:
(a) storing larger functions in their own files (e.g. ~/myApp/R/function-name.R) and / or
(b) creating a utils.R file (e.g ~/myApp/R/utils.R) to store smaller, simpler functions all in one script
You can source your function files into global.R so that your functions are made available for use throughout your app.
NOTE: As of Shiny version 1.5.0, any scripts stored in ~/myApp/R/ will be automatically sourced when your application is loaded (meaning you don’t need to source() them into global.R, if you’re running at least Shiny v1.5.0).
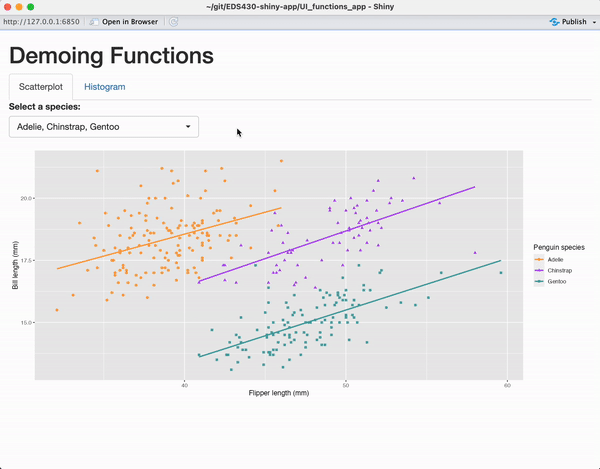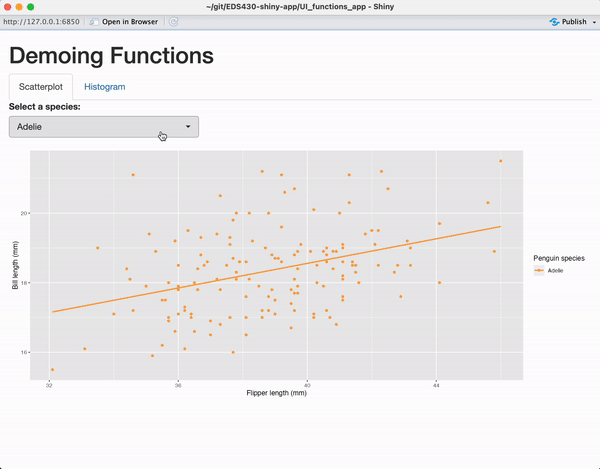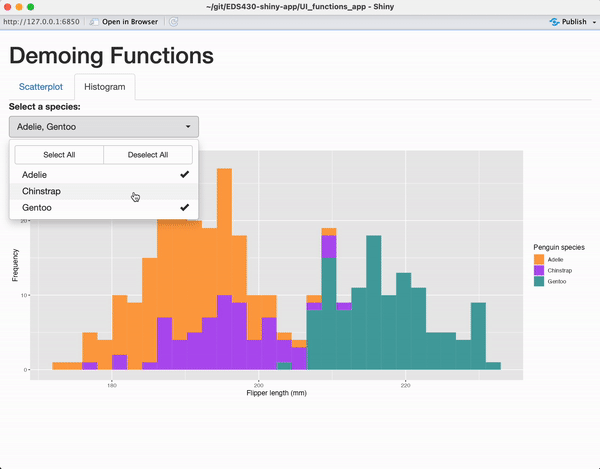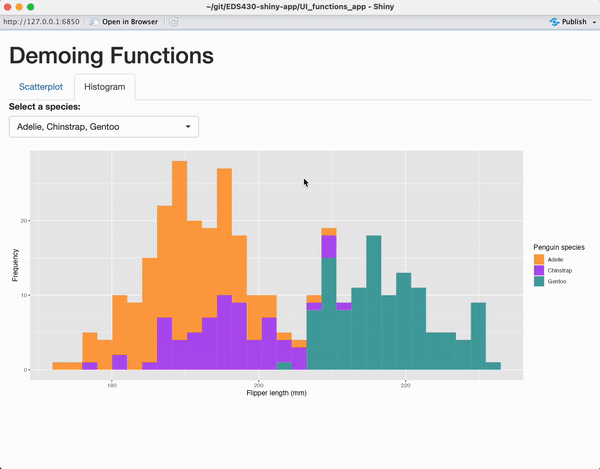
Create a small app for function practice
Add the following files (+ code) to a new subdirectory called ~/functions-app/, and check out the resulting app:

~/functions-app/ui.R
ui <- fluidPage(
tags$h1("Demoing Functions"),
# tabsetPanel ----
tabsetPanel(
# scatterplot tab ----
tabPanel(title = "Scatterplot",
# species (scatterplot) pickerInput ----
pickerInput(inputId = "penguin_spp_scatterplot_input",
label = "Select a species:",
choices = c("Adelie", "Chinstrap", "Gentoo"),
selected = c("Adelie", "Chinstrap", "Gentoo"),
options = pickerOptions(actionsBox = TRUE),
multiple = TRUE),
# scatterplot output ----
plotOutput(outputId = "penguin_scatterplot_output")
), # END scatterplot tab
# histogram tab ----
tabPanel(title = "Histogram",
# species (histogram) pickerInput ----
pickerInput(inputId = "penguin_spp_histogram_input",
label = "Select a species:",
choices = c("Adelie", "Chinstrap", "Gentoo"),
selected = c("Adelie", "Chinstrap", "Gentoo"),
options = pickerOptions(actionsBox = TRUE),
multiple = TRUE),
# scatterplot output ----
plotOutput(outputId = "penguin_histogram_output")
) # END histogram tab
) # END tabsetPanel
) # END fluidPage~/functions-app/server.R
server <- function(input, output) {
# filter penguin species (scatterplot) ----
filtered_spp_scatterplot_df <- reactive ({
penguins |>
filter(species %in% input$penguin_spp_scatterplot_input)
})
# render the scatterplot output ----
output$penguin_scatterplot_output <- renderPlot({
ggplot(na.omit(filtered_spp_scatterplot_df()),
aes(x = bill_length_mm, y = bill_depth_mm,
color = species, shape = species)) +
geom_point() +
geom_smooth(method = "lm", se = FALSE, aes(color = species)) +
scale_color_manual(values = c("Adelie" = "darkorange",
"Chinstrap" = "purple",
"Gentoo" = "cyan4")) +
scale_shape_manual(values = c("Adelie" = 19,
"Chinstrap" = 17,
"Gentoo" = 15)) +
labs(x = "Flipper length (mm)",
y = "Bill length (mm)",
color = "Penguin species",
shape = "Penguin species")
})
# filter penguin species (histogram) ----
filtered_spp_histogram_df <- reactive ({
penguins |>
filter(species %in% input$penguin_spp_histogram_input)
})
# render the histogram output ----
output$penguin_histogram_output <- renderPlot({
ggplot(na.omit(filtered_spp_histogram_df()),
aes(x = flipper_length_mm, fill = species)) +
geom_histogram(alpha = 0.5, position = "identity") +
scale_fill_manual(values = c("Adelie" = "darkorange",
"Chinstrap" = "purple",
"Gentoo" = "cyan4")) +
labs(x = "Flipper length (mm)",
y = "Frequency",
fill = "Penguin species")
})
} # END serverIdentify code duplication in ui.R
Let’s first focus on the UI – where do we have nearly identically duplicated code?
~/functions-app/ui.R
ui <- fluidPage(
tags$h1("Demoing Functions"),
# tabsetPanel ----
tabsetPanel(
# scatterplot tab ----
tabPanel(title = "Scatterplot",
# species (scatterplot) pickerInput ----
pickerInput(inputId = "penguin_spp_scatterplot_input",
label = "Select a species:",
choices = c("Adelie", "Chinstrap", "Gentoo"),
selected = c("Adelie", "Chinstrap", "Gentoo"),
options = pickerOptions(actionsBox = TRUE),
multiple = TRUE),
# scatterplot output ----
plotOutput(outputId = "penguin_scatterplot_output")
), # END scatterplot tab
# histogram tab ----
tabPanel(title = "Histogram",
# species (histogram) pickerInput ----
pickerInput(inputId = "penguin_spp_histogram_input",
label = "Select a species:",
choices = c("Adelie", "Chinstrap", "Gentoo"),
selected = c("Adelie", "Chinstrap", "Gentoo"),
options = pickerOptions(actionsBox = TRUE),
multiple = TRUE),
# scatterplot output ----
plotOutput(outputId = "penguin_histogram_output")
) # END histogram tab
) # END tabsetPanel
) # END fluidPageWe can turn these pickerInputs into a function
This app includes two pickerInputs, both of which allow users to select which penguin species to display data for. The only difference between both pickerInputs is the inputId.
# Scatterplot pickerInput for selecting penguin species:
pickerInput(inputId = "penguin_spp_scatterplot_input",
label = "Select a species:",
choices = c("Adelie", "Chinstrap", "Gentoo"),
selected = c("Adelie", "Chinstrap", "Gentoo"),
options = pickerOptions(actionsBox = TRUE),
multiple = TRUE)
# Histogram pickerInput for selecting penguin species:
pickerInput(inputId = "penguin_spp_histogram_input",
label = "Select a species:",
choices = c("Adelie", "Chinstrap", "Gentoo"),
selected = c("Adelie", "Chinstrap", "Gentoo"),
options = pickerOptions(actionsBox = TRUE),
multiple = TRUE)Let’s write a function for our penguin species pickerInput that we can use in place of these two, rather long, chunks of code.
Write a function for adding a pickerInput to select for penguin species
First, create an R/ folder inside your functions-app/ directory, then add a new script to this folder. I’m calling mine penguinSpp-pickerInput.R.
Since the only difference between our original two pickerInputs are their inputIds, we can write a function that takes inputId as an argument (Recall that inputIds must be unique within an app, so it makes sense that both of our pickerInputs have different inputIds).
Once written, source() your function script into global.R (if necessary) to make your function available for use in your app.
~/functions-app/R/penguin_spp-pickerInput.R
Apply your function in ui.R
Finally, replace your original UI code for building both pickerInputs with our penguin_spp_pickerInput() function, save, and run your app. It should look exactly the same as before!
~/functions-app/ui.R
ui <- fluidPage(
tags$h1("Demoing Functions"),
# tabsetPanel ----
tabsetPanel(
# scatterplot tab ----
tabPanel(title = "Scatterplot",
# species (scatterplot) pickerInput ----
penguin_spp_pickerInput(inputId = "penguin_spp_scatterplot_input"),
# scatterplot output ----
plotOutput(outputId = "penguin_scatterplot_output")
), # END scatterplot tab
# histogram tab ----
tabPanel(title = "Histogram",
# species (histogram) pickerInput ----
penguin_spp_pickerInput(inputId = "penguin_spp_histogram_input"),
# scatterplot output ----
plotOutput(outputId = "penguin_histogram_output")
) # END histogram tab
) # END tabsetPanel
) # END fluidPageWe reduced code redundancy and increased readability!
So…what’s the big deal with this??
By turning our pickerInput code into a function, we:
(1) reduced ten lines of UI code into two (not only does this make ui.R a bit more manageable to navigate, it also means we can more easily isolate R/penguinSpp-pickerInput.R when troubleshooting)
(2) made our UI code a bit easier to read (penguin_spp_pickerInput() tells a reader / collaborator / future you exactly what that line of code is meant to do, which is to create a pickerInput that allows users to select penguin species. Even without code comments or additional context, one may deduce what that line of code does)
Identify where we can streamline our server
Next, let’s see where we can streamline our server code using functions. We have two discrete sections of code – (1) a reactive data frame and scatterplot output and (2) a reactive data frame and histogram output.
~/functions-app/server.R
server <- function(input, output) {
# filter penguin species (scatterplot) ----
filtered_spp_scatterplot_df <- reactive ({
penguins |>
filter(species %in% input$penguin_spp_scatterplot_input)
})
# render the scatterplot output ----
output$penguin_scatterplot_output <- renderPlot({
ggplot(na.omit(filtered_spp_scatterplot_df()),
aes(x = bill_length_mm, y = bill_depth_mm,
color = species, shape = species)) +
geom_point() +
geom_smooth(method = "lm", se = FALSE, aes(color = species)) +
scale_color_manual(values = c("Adelie" = "#FEA346",
"Chinstrap" = "#B251F1",
"Gentoo" = "#4BA4A4")) +
scale_shape_manual(values = c("Adelie" = 19,
"Chinstrap" = 17,
"Gentoo" = 15)) +
labs(x = "Flipper length (mm)",
y = "Bill length (mm)",
color = "Penguin species",
shape = "Penguin species")
})
# filter penguin species (histogram) ----
filtered_spp_histogram_df <- reactive ({
penguins |>
filter(species %in% input$penguin_spp_histogram_input)
})
# render the histogram output ----
output$penguin_histogram_output <- renderPlot({
ggplot(na.omit(filtered_spp_histogram_df()),
aes(x = flipper_length_mm, fill = species)) +
geom_histogram(alpha = 0.5, position = "identity") +
scale_fill_manual(values = c("Adelie" = "#FEA346",
"Chinstrap" = "#B251F1",
"Gentoo" = "#4BA4A4")) +
labs(x = "Flipper length (mm)",
y = "Frequency",
fill = "Penguin species")
})
} # END serverCreate a function to build our scatterplot
The goal of this function is to filter the penguins data based on the user input and render our ggplot scatterplot. To start, I’m going to cut / paste both the code to generate the reactive filtered_spp_scatterplot_df data frame and the renderPlot() code from server.R into our build_penguin_scatterplot() function.
~/functions-app/R/build-penguin-scatterplot.R
build_penguin_scatterplot <- function(input) {
# filter penguin species (scatterplot) ----
filtered_spp_scatterplot_df <- reactive ({
penguins |>
filter(species %in% input$penguin_spp_scatterplot_input)
})
# render the scatterplot output ----
renderPlot({
ggplot(na.omit(filtered_spp_scatterplot_df()),
aes(x = bill_length_mm, y = bill_depth_mm,
color = species, shape = species)) +
geom_point() +
geom_smooth(method = "lm", se = FALSE, aes(color = species)) +
scale_color_manual(values = c("Adelie" = "#FEA346",
"Chinstrap" = "#B251F1",
"Gentoo" = "#4BA4A4")) +
scale_shape_manual(values = c("Adelie" = 19,
"Chinstrap" = 17,
"Gentoo" = 15)) +
labs(x = "Flipper length (mm)",
y = "Bill length (mm)",
color = "Penguin species",
shape = "Penguin species")
})
}A note on the function argument, input
Important: In isolation, our function does not know about the user input (input is not in our global environment, it’s only known within the server() function). Therefore, we must pass input as an argument to our function. This makes any user-supplied inputs from the UI available to our function, build_penguin_scatterplot(), so that we can successfully filter the penguins data.
build_penguin_scatterplot() will return the object created by renderPlot() (i.e. our rendered scatterplot).
Now use your function inside the server
Remember, the output of build_penguin_scatterplot() is renderPlot(), which is used to build our reactive scatterplot. Following our rules for creating reactivity, we need to save our function’s output to output$penguin_scatterplot_output. In doing so, we reduced 23 lines of code to 1 inside our server function.
~/functions-app/server.R
server <- function(input, output) {
# filter data & create penguin scatterplot ----
output$penguin_scatterplot_output <- build_penguin_scatterplot(input)
# filter penguin species (histogram) ----
filtered_spp_histogram_df <- reactive ({
penguins |>
filter(species %in% input$penguin_spp_histogram_input)
})
# render the histogram output ----
output$penguin_histogram_output <- renderPlot({
ggplot(na.omit(filtered_spp_histogram_df()),
aes(x = flipper_length_mm, fill = species)) +
geom_histogram(alpha = 0.5, position = "identity") +
scale_fill_manual(values = c("Adelie" = "#FEA346",
"Chinstrap" = "#B251F1",
"Gentoo" = "#4BA4A4")) +
labs(x = "Flipper length (mm)",
y = "Frequency",
fill = "Penguin species")
})
} # END serverBuild a function to create our histogram
We can repeat a similar process to create a function for building our histogram:
~/functions-app/R/build-penguin-histogram.R
build_penguin_histogram <- function(input) {
# filter penguin spp ----
filtered_spp_histogram_df <- reactive ({
penguins |>
filter(species %in% input$penguin_spp_histogram_input)
})
# render histogram ----
renderPlot({
ggplot(na.omit(filtered_spp_histogram_df()),
aes(x = flipper_length_mm, fill = species)) +
geom_histogram(alpha = 0.5, position = "identity") +
scale_fill_manual(values = c("Adelie" = "#FEA346",
"Chinstrap" = "#B251F1",
"Gentoo" = "#4BA4A4")) +
labs(x = "Flipper length (mm)",
y = "Frequency",
fill = "Penguin species")
})
}~/functions-app/server.R
Final code (1/3)
Run your updated app to ensure it works as expected. Your final code should look like this:
~/functions-app/ui.R
ui <- fluidPage(
tags$h1("Demoing Functions"),
# tabsetPanel ----
tabsetPanel(
# scatterplot tab ----
tabPanel(title = "Scatterplot",
# species (scatterplot) pickerInput ----
penguin_spp_pickerInput(inputId = "penguin_spp_scatterplot_input"),
# scatterplot output ----
plotOutput(outputId = "penguin_scatterplot_output")
), # END scatterplot tab
# histogram tab ----
tabPanel(title = "Histogram",
# species (histogram) pickerInput ----
penguin_spp_pickerInput(inputId = "penguin_spp_histogram_input"),
# scatterplot output ----
plotOutput(outputId = "penguin_histogram_output")
) # END histogram tab
) # END tabsetPanel
) # END fluidPage~/functions-app/server.R
Final code (2/3)
penguinSpp-pickerInput.R
~/functions-app/R/penguin_spp-pickerInput.R
Final code (3/3)
~/functions-app/R/build-penguin-scatterplot.R
build_penguin_scatterplot <- function(input) {
# filter penguin species (scatterplot) ----
filtered_spp_scatterplot_df <- reactive ({
penguins |>
filter(species %in% input$penguin_spp_scatterplot_input)
})
# render the scatterplot output ----
renderPlot({
ggplot(na.omit(filtered_spp_scatterplot_df()),
aes(x = bill_length_mm, y = bill_depth_mm,
color = species, shape = species)) +
geom_point() +
geom_smooth(method = "lm", se = FALSE, aes(color = species)) +
scale_color_manual(values = c("Adelie" = "#FEA346",
"Chinstrap" = "#B251F1",
"Gentoo" = "#4BA4A4")) +
scale_shape_manual(values = c("Adelie" = 19,
"Chinstrap" = 17,
"Gentoo" = 15)) +
labs(x = "Flipper length (mm)",
y = "Bill length (mm)",
color = "Penguin species",
shape = "Penguin species")
})
}~/functions-app/R/build-penguin-histogram.R
build_penguin_histogram <- function(input) {
# filter penguin spp ----
filtered_spp_histogram_df <- reactive ({
penguins |>
filter(species %in% input$penguin_spp_histogram_input)
})
# render histogram ----
renderPlot({
ggplot(na.omit(filtered_spp_histogram_df()),
aes(x = flipper_length_mm, fill = species)) +
geom_histogram(alpha = 0.5, position = "identity") +
scale_fill_manual(values = c("Adelie" = "#FEA346",
"Chinstrap" = "#B251F1",
"Gentoo" = "#4BA4A4")) +
labs(x = "Flipper length (mm)",
y = "Frequency",
fill = "Penguin species")
})
}End part 6.1
Up next: modules
05:00